New Launch – Mycoplasma qPCR Detection Kit (Fluorescent Probe Method)
I.Background and Application
the research and production of cell therapy products, pathogen contamination may pose significant clinical risks. Traditional detection methods, such as culture-based assays, are limited by long turnaround times and insufficient sensitivity. Quantitative PCR (qPCR), with its high sensitivity, rapid turnaround, and strong specificity, has become a critical technology for quality control in cell therapy. This kit has been specifically developed to address the unique requirements of cell therapy products. It enables accurate detection of a wide range of pathogen contaminants, ensuring safety throughout the entire process—from cell bank establishment to final product release—thus providing reliable assurance for compliant manufacturing of cell therapy products.
This product series is based on the TaqMan probe assay, enabling both qualitative and quantitative detection of target pathogen nucleic acids. It is suitable for applications in cell therapy, biopharmaceutical manufacturing, and scientific research. Covering the entire workflow from cell banking, culture monitoring, to final product release, it provides precise detection of microbial contaminants (bacteria, fungi, mycoplasma, and viruses). The kit complies with pharmacopeial standards and regulatory requirements, ensuring reliable safety control for cell therapy.
II.Product List
| Catalog No. | Product Name |
| MP-D050T/MP-D100T | Mycoplasma qPCR Detection Kit (Fluorescent Probe Method) |
III.Principle of Assay
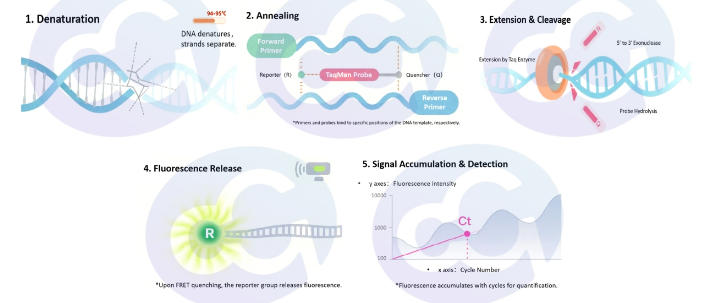
The TaqMan probe assay is a fluorescence-based quantitative PCR (qPCR) detection technology. It utilizes specific primers and a fluorescently labeled probe that binds to the target nucleic acid. The probe is labeled with a fluorescent reporter (R) at the 5’ end and a quencher (Q) at the 3’ end. When the probe is intact, the fluorescence emitted by the reporter is absorbed by the quencher, resulting in no detectable signal. During qPCR amplification, the Taq polymerase cleaves the probe, separating the reporter from the quencher and generating a fluorescent signal. The intensity of the fluorescence is proportional to the amount of target nucleic acid, enabling highly specific and sensitive quantitative analysis.
IV.Advantages of Kit
Highly Sensitive Detection: Sensitivity reaches 1–10 copies/μL, enabling identification of extremely low-level contamination risks.
Accurate Discrimination: Probes specifically recognize target sequences, minimizing false positives and false negatives.
End-to-End Monitoring: Covers the entire workflow from cell bank establishment and culture monitoring to final product release.
Wide Applicability: Suitable for cell therapy, biopharmaceutical production, and research applications.
Platform Compatibility: Compatible with qPCR instruments from various manufacturers and models.
Compliance and Reliability: Fully compliant with ChP, EP, USP, and other pharmacopeial standards.
V.Performance Specifications
Complies with mycoplasma detection requirements outlined in the European Pharmacopoeia (EP 2.6.7), United States Pharmacopeia (USP 63), and Japanese Pharmacopoeia (JP 18).
a)Sensitivity and Detection Limit
The kit achieves a sensitivity of at least 10 CFU/mL, with a detection rate of ≥95%.
Sample | Strain Source | Concentration | Positive Results / Total Tests |
Oral mycoplasma | ATCC 23714 | 10 CFU/mL | 23/24 |
Mycoplasma pneumoniae | ATCC 15531 | 10 CFU/mL | 24/24 |
Mycoplasma hyorhinis | CVCC 361 | 10 CFU/mL | 24/24 |
b)Specificity
Capable of specifically detecting over 100 mycoplasma species, with no cross-reactivity observed for the following bacteria, fungi, host cell genomic DNA, or culture media. No Ct values are generated for these non-targets.
| No. | Bacterial Strain | No. | Cell Line | No. | Solution / Medium |
| 1 | Escherichia coli | 8 | MDCK | 14 | PBS |
| 2 | Klebsiella pneumoniae | 9 | 293T | 15 | FBS |
| 3 | Streptococcus pneumoniae | 10 | CHO-K1 | 16 | RPMI-1640 medium |
| 4 | Staphylococcus aureus | 11 | vero | 17 | MEM medium |
| 5 | Clostridium difficile | 12 | Hela | 18 | CHO medium |
| 6 | Lactobacillus acidophilus | 13 | BHK-21 | 19 | L-G medium |
| 7 | Pichia pastoris | 20 | PS medium |
c)Matrix Effect
The addition of mycoplasma standards into the following matrices did not affect the assay results or overall kit performance.
| No. | Sample |
| 1 | 293T cells |
| 2 | 293T cell culture supernatant |
| 3 | CHO-K1 cells |
| 4 | CHO-K1 cell culture supernatant |
| 5 | Vero cells |
| 6 | Vero cell culture supernatant |
| 7 | RPMI1640 medium |
| 8 | DMEM medium |
| 9 | 10% FBS |
d)Freeze–Thaw Stability
The kit was subjected to repeated freeze-thaw cycles (1-5 times), and no impact on assay results or overall performance was observed
e)Repeatability
The intra-assay and inter-assay coefficients of variation (CV) were both less than 10%, demonstrating good repeatability.
f)Compatibility
When tested on different real-time PCR instruments (ABI 7500, Bioer FQD-96A, and BIO-RAD CFX96) using the same samples, the kit accurately identified mycoplasma-positive and -negative results. The ABI 7500 showed higher sensitivity compared with the other two instruments.
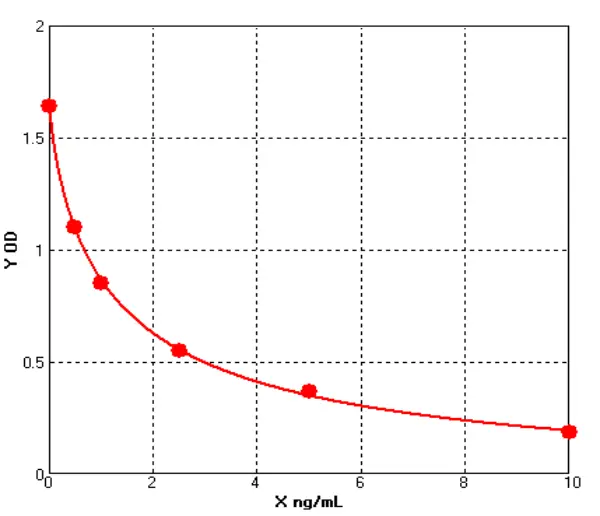
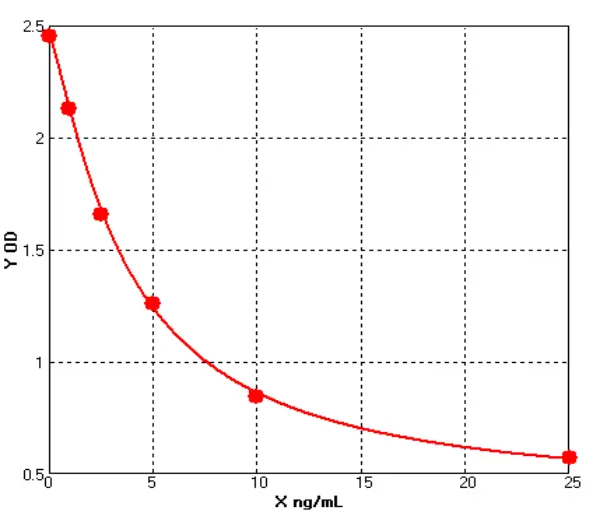
VI.Test Results (Example: Mycoplasma)
1. Amplification Curve
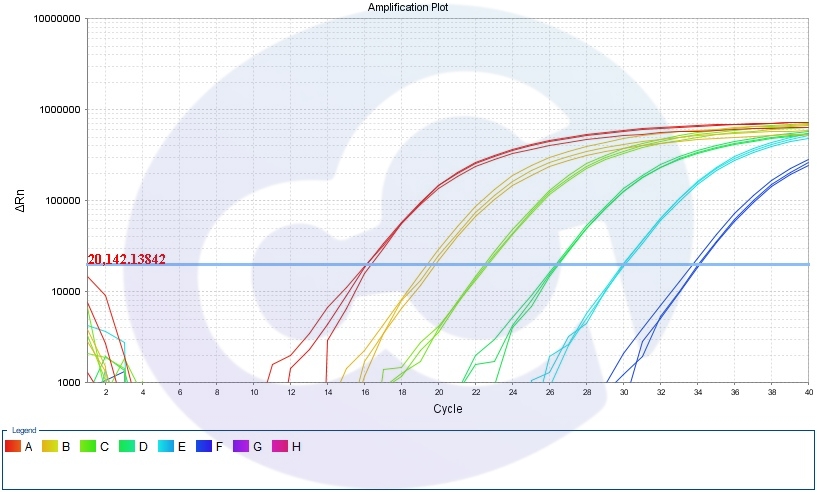
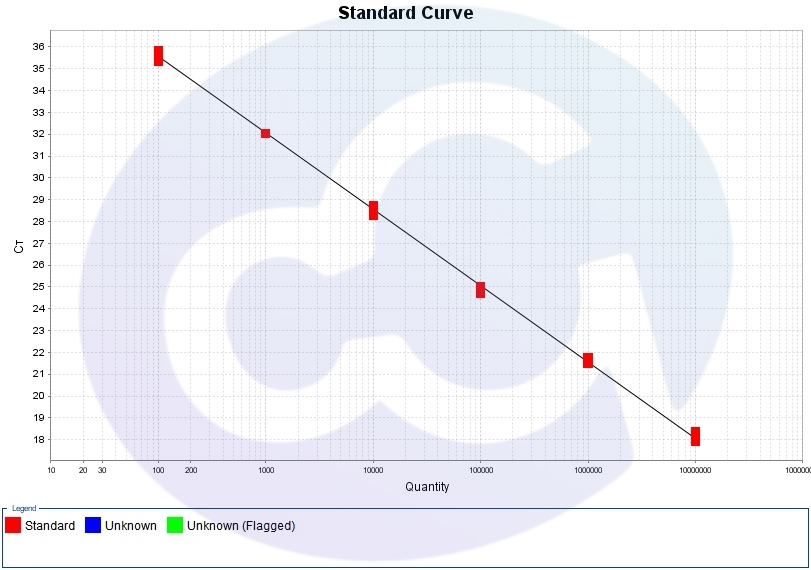
2.Standard Curve
![]()
Parameter | Standard | |
Standard Curve | Slope | -3.1~-3.8 |
Linearity(R2) | >0.99 | |
Amplification efficiency (Eff) | 90%-110% | |
Quality Control | Sensitivity | 10 CFU |
Coefficient of Variation (CV) | ≤10% | |
Specificity | √ | |
Exclusivity | √ | |
Robustness | √ |